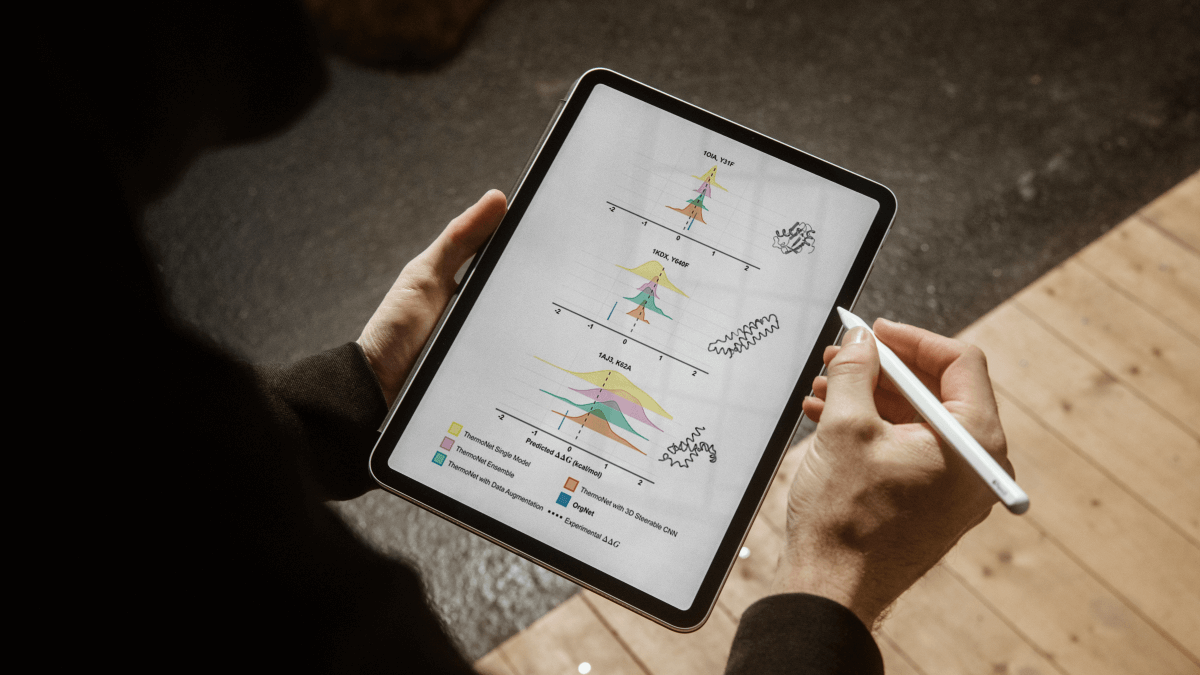
Accurately predicting how single-point mutations affect protein stability is vital for understanding genetic diseases and designing new proteins for biotechnology and medicine. Traditional computational methods have struggled with accuracy and robustness, thus, novel approaches are in great need. 3D convolutional neural networks proved to be powerful, but are not robust with respect to the input orientations.
OrgNet solves the orientation bias problem by introducing a novel orientation-gnostic 3D convolutional neural network, which standardizes protein structures before analysis. This eliminates orientation bias and allows the model to capture fine-grained atomic details more reliably.
The model has implications for both biomedicine and biotechnology. OrgNet can help identify mutations that destabilize proteins, shedding light on the molecular basis of diseases. In biotechnology, it offers a powerful tool for engineering proteins with desired stability properties, accelerating industrial enzyme development and synthetic biology applications.
The study was recently published in Bioinformatics and presented at ISMB/ECCB 2025, one of the most recognized conferences in computational biology. Read the full article here.